A ONE-STOP BIOINFORMATICS PLATFORM
A one-stop bioinformatics platform is in development phase, which aims to support a variety of genomics, transcriptomics, proteomics, metagenomics and other next generation sequencing analysis. As a part of this platform, a central data repository will be maintained within HPC, with all the needed datasets like different versions of genome references (human, mouse, fly etc), indexed genomes for different aligners (bowtie1/2, STAR & other aligner indexes) and commonly used reference databases like NCBI (nr, est etc.). This central repository helps users to make use of their user space more for their downstream analysis. Users can also request for any specific data to be available inside the central repository.
Nextflow pipelines
As a part of this platform, we are developing few of the best practised pipelines using nextflow for easy implementation. Nextflow is efficient for reproducibility, and simplifies the execution of complex distributed computational workflows in a portable and reproducible manner. It allows seamless parallelisation and tight integration with existing HPC’s job scheduler. Some existing pipelines include RNA-seq which does the Quality Control (using FastQC, fastx-toolkit), Alignment (bowtie/STAR) and Differential expression (Cufflinks/rsem) and other pipelines are in development phase.
A simple blast pipeline on HPC
Some example pipelines are made available for user to work on nextflow. Here we demonstrate a simple blast pipeline which runs in parallel for each fasta sequence using a parallel8 job queue. The first blast process implements a blast query using blastp and saves the top 10 results. The next extract process takes the output of blast process as input and finds the top 10 matches returned by the blast query. Several such processes can be linked by redirecting the output of one process as input to another. Such linked processes will always get executed in order.
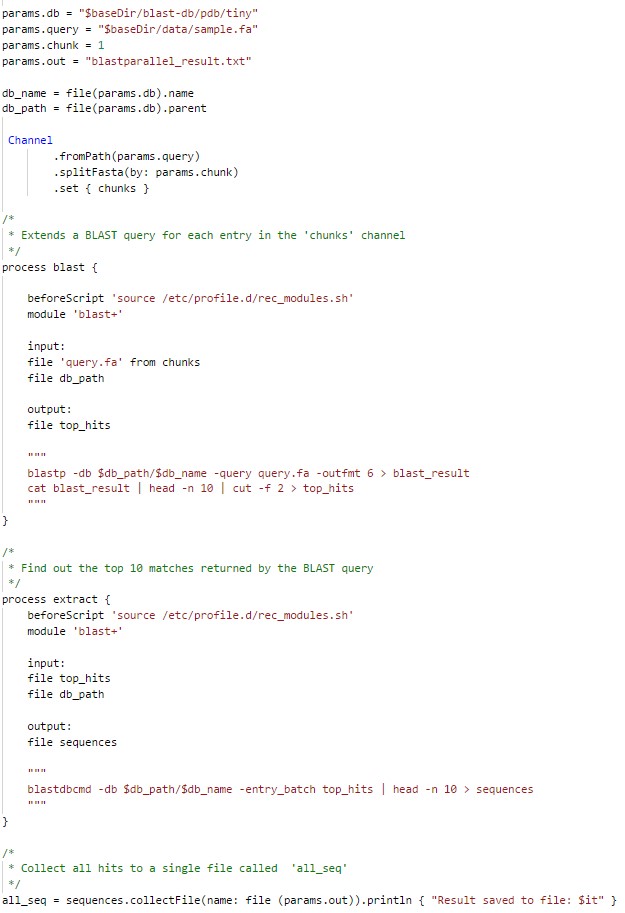
In the above example, an input channel is created with each fasta record as an entry within the channel. The blast process will be first run in parallel on each fasta record and once all the blast processes are executed, nextflow will run extract process using the output of blast process as input. At the end, results of extract processes are collected into a single output file. As the nextflow is configured to on hpc queues, you need to choose the appropriate profiles while executing nextflow jobs.
By default nextflow, runs the jobs in local cluster/system. You should choose p8/p12/p24 profile, to submit the jobs to parallel8/parallel12/parallel24 queue respectively. These profiles are configured inside the nextflow.config file that is present in the same execution directory.
>$ nextflow run blast-parallel.nf (#executed in local mode) >$ nextflow run blast-parllel.nf -profile p8 (# for submitting to parallel8 queue) N E X T F L O W ~ version 0.26.3 Launching `blast-parallel.nf` [exotic_mccarthy] - revision: f9aa995d38 [warm up] executor > pbs [50/da3566] Submitted process > blast (1) [4a/d30c5b] Submitted process > blast (2) [c7/961755] Submitted process > blast (4) [30/053cbf] Submitted process > blast (5) [44/32716d] Submitted process > blast (3) [84/538d65] Submitted process > extract (1) [6b/17bbba] Submitted process > extract (2) [5b/4c807c] Submitted process > extract (3) [e5/ddf8d8] Submitted process > extract (4) [6c/630f4d] Submitted process > extract (5) Result saved to file: blastparallel_result.txt
These pipelines are useful for reproducing the same analysis in any computing environment (local/hpc cluster/cloud).
Using Bioinformatics pipelines
Researchers can either make use of the existing best practised pipelines or develop their own nextflow pipelines using existing pipeline templates. Users can also reach us at DataEngineering@nus.edu.sg for more information or any assistance in pipeline development.

