FACILITATING COMPUTER AIDED DRUG DISCOVERY BY GPU ACCELERATES MOLECULAR DYNAMICS SIMULATIONS
Computer aided drug discovery (CADD) 1 is now extensively implemented in the early stage of drug design/discovery projects. Structure-based virtual screening is one of the commonly used CADD approaches. In this approach, docking calculations are performed to filter entries from huge compound libraries against a target protein with known 3D structure. Due to the remarkable advancement in computer facility and robust molecular docking software, thousands of compounds can be screened within a few days so that the efforts and costs of subsequent experimental works are greatly reduced. In our study, structure-based virtual screening is applied to find novel compounds that can bind to and stabilize our protein of interest, RARα.
Our recent study revealed that the RARA gene is frequently mutated in breast fibroepithelial tumors and suggested a role of RAR signaling in tumorigenesis2. RARA gene encodes RARα protein, a nuclear receptor that regulates the expression of target genes by selective recruitment of co-activator or corepressor complexes. In the presence of an agonist (e.g. retinoic acid (RA)), RARα recruits co-activators, resulting in transcriptional activation of target genes. In the absence of an agonist, RARα preferentially binds to co-repressors, giving rise to the constitutive repression of target gene expressions.
Besides RA, classes of synthetic ligands have been developed and studied. For instance, neutral antagonists such BMS614 prevents the recruitment of co-activator whereas inverse agonist such as BMS493 strengthens the interactions between RARα and co-repressor. In our previous study, we found the breast fibroepithelial tumor-associated RARα mutations to be all located in the ligandbinding domain (LBD). In vitro studies it showed that these RARα mutants lost the sensitivity to RA treatment2. Taken together, these results implied that the alteration of binding modes and affinities of agonists might occur in mutant RARα conformations. To find novel compounds that can bind RARα and recover its co-activator recruiting activity, we adopted relaxed complex scheme (RCS) to screen compounds from the compound library, ZINC database3.
The flexible receptor usually gives better accuracy than the static receptor when docking small molecules to a protein receptor. Based on this idea, the design of RCS docking scheme is to dock compounds to multiple protein conformations that introduces receptor flexibility to docking calculations4. Protein conformations can be obtained by molecular dynamics (MD) simulations or by other equivalent conformational ensemble generating methods such as Monte Carlo simulations, Brownian dynamics simulations, etc. In our case, the X-ray crystal structures of RARα LBD are available in Protein Data Bank5. There are five published RARα LBD structures in complex with either agonists or antagonists (neutral antagonist and inverse agonist). We chose RA-bound conformation (PDB ID: 3A9E6) as the template to build the structural models of mutant proteins and then performed MD simulations for the wild-type and mutant models.
In our system, the protein was immersed in a TIP3P water box. Counter-ions (Na+ and Cl-) were added to mimic physiological ion concentration (0.15M) and to make the system electrostatic neutrally as well. Figure 1 demonstrates the system setup of the wild-type RARα as an example. There are 29197 atoms in the simulation box containing RARα LBD, RA, water molecules and counter-ions. The system was first energy minimized for 2000 steps and then gradually heated up to 298.15K within 50ps. For the production of conformational ensemble, we applied GPU version of PMEMD provided by AMBER 14 package7. Before the routine submission of production jobs, we did a performance test for the comparison of CPU and GPU versions of PMEMD. The benchmark showed that GPU significantly boosted the calculation speed. In Table 1, GPU computational scheme gave more than 13 fold speedup of the calculation. For our system of roughly 30000 atoms, it took about one week to simulate 100ns-long protein dynamics in an explicit solvent environment.
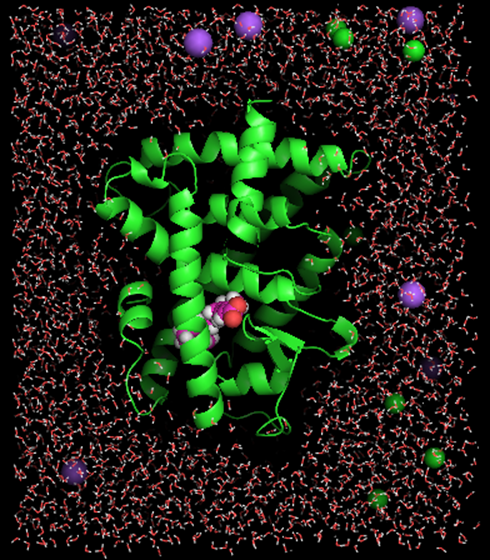
For each mutant RARα model, we plan to perform 500ns long dynamics simulations. The obtained protein conformations will be grouped according to their root-mean-square deviation (RMSD) values by clustering algorithm and the representative conformations of each cluster will be used as receptors in virtual screening of a compound library.
Table 1. Comparison of performance between CPU and GPU computational schemes
| Scheme | CPU spec | GPU spec | Program | ns/day | Speedup |
|---|---|---|---|---|---|
| 8 x CPU | 8Xeon X5650 2.67 GHz |
N/A | pmemd.MPI | 1.40 | 1.00 |
| 8 x CPU 2 x GPU |
Xeon X5650 2.67 GHz |
Tesla M2090 1.30 GHz |
pmemd.cuda_SPFP.MPI | 18.29 | 13.06 |
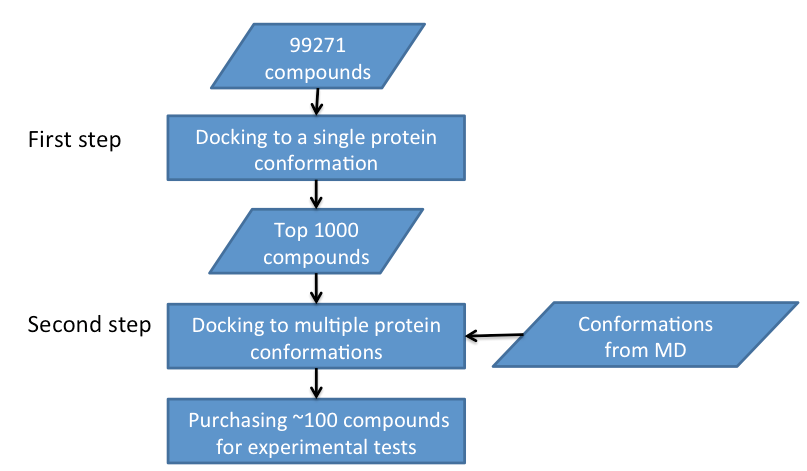
The screening protocol is shown in Figure 2. Due to the limited computational resource, the screening work is split into two steps. In the first step, compounds will be dock to a single protein conformation. Only the top 1000 compound candidates will be further screened by RCS docking scheme. The selected compounds will be dock to multiple protein conformations generated by MD simulations and clustering. Finally, top 100 compounds will be experimentally tested to validate their in vitro activities.
References
- Sliwoski G, Kothiwale S, Meiler J, Lowe EW, Jr. Computational methods in drug discovery.
Pharmacol Rev 66, 334-395 (2014). - Tan J, et al. Genomic landscapes of breast fibroepithelial tumors.
Nat Genet 47, 1341-1345 (2015). - Irwin JJ, Sterling T, Mysinger MM, Bolstad ES, Coleman RG. ZINC: a free tool to discover chemistry for biology.
J Chem Inf Model 52, 1757-1768 (2012). - Lin JH, Perryman AL, Schames JR, McCammon JA. Computational drug design accommodating receptor flexibility: the relaxed complex scheme.
J Am Chem Soc 124, 5632-5633 (2002). - Berman HM, et al. The Protein Data Bank.
Nucleic Acids Res 28, 235-242 (2000). - Sato Y, et al. The “Phantom Effect” of the Rexinoid LG100754: structural and functional insights.
PLoS One 5, e15119 (2010). - Salomon-Ferrer R, Gotz AW, Poole D, Le Grand S, Walker RC. Routine Microsecond Molecular Dynamics Simulations with AMBER on GPUs. 2. Explicit Solvent Particle Mesh Ewald.
J Chem Theory Comput 9, 3878-3888 (2013). - Kannan S, Ganji R. Porting Autodock to CUDA.
In: IEEE Congress on Evolutionary Computation (2010). - Ouyang X, Kwoh CK. GPU Accelerated Molecular Docking with Parallel Genetic Algorithm.
In: Parallel and Distributed Systems (ICPADS), 2012
IEEE 18th International Conference on (2012).

