ACCELERATION OF MOLECULAR DYNAMICS SIMULATION USING AMBER GPU VERSION
With the explosive growth of the number of GPU accelerated application in the last one to two years, General Purpose GPU (GP-GPU) have started to play a more important role in scientific computing. For Molecular Dynamics (MD) simulation applications, most of them have GPU versions now and it is discovered that these MD code runs faster on a GPU-CPU system than on CPU only systems. Because of this progress, scientist can now run simulations on larger biological systems with longer timeframes, which means, instead of nanosecond simulation, microsecond MD simulations have become common now.
How does GPU works?
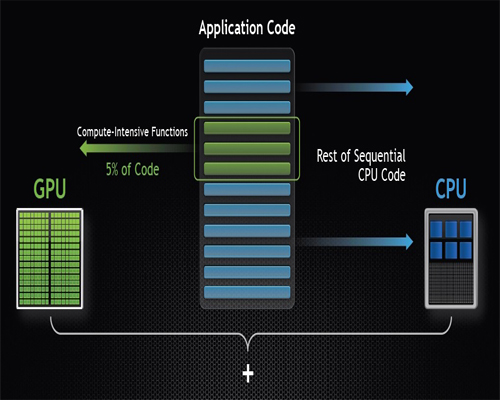
Normally GPU works together with CPUs to accelerate scientific, engineering, and enterprise applications. As shown in Figure 1, GPU-accelerated computing offers application performance by offloading compute-intensive portions of the application to the GPU, while the remainder of the code still runs on the CPU. A GPU consists of thousands of smaller, more efficient cores designed for handling multiple tasks simultaneously. Hence, it works best for data-parallel computation in a SIMD (Single Instruction Multiple Data) kind of parallelism.
AMBER GPU Benchmark
Amber is a popular software package for biomolecular simulations. Starting from Amber11, several key features are introduced in the GPU version. These features were further extended in AMBER 12 and the performance envelope has been pushed even further with AMBER 14.
The following benchmark was carried out using the Amber 12 PMEMD module, which is an extensively-modified version of the Amber sander program, optimized for periodic, PME (particle-mesh Ewald) simulations, and for GB (Generalized Born) simulations.
Different CPU and GPUs are used for the benchmark execution on a compute node with two Intel X5650 6-core processors at 2.66 GHz and 48 GB RAM, and two NVIDIA Tesla M2090 512-cores processors at 650 MHz and 6 GB memory.
The results are given in the following table:
| JOBID | EXECUTABLES | No. of CPUs | No. of GPU | Elapsed (s) | ns/day | average(ns/day) |
|---|---|---|---|---|---|---|
| 782824 | pmpd.mpi | 12 | 0 | 17,075.47 | 2.53 | 2.20 |
| 783462 | pmpd.mpi | 12 | 0 | 26,083.70 | 1.66 | |
| 788682 | pmpd.mpi | 12 | 0 | 17,881.63 | 2.42 | |
| 813594 | pmpd.cuda | 1 | 1 | 4,108.01 | 10.52 | 10.55 |
| 813599 | pmpd.cuda | 1 | 1 | 4,128.73 | 10.46 | |
| 870498 | pmpd.cuda | 1 | 1 | 4,045.63 | 10.68 | |
| 782218 | pmpd.cuda.mpi | 2 | 2 | 3,007.33 | 14.25 | 14.11 |
| 782487 | pmpd.cuda.mpi | 2 | 2 | 3,120.33 | 13.73 | |
| 820474 | pmpd.cuda.mpi | 2 | 2 | 3,007.58 | 14.36 |
Table 1: Amber benchmark results based on CPUs and CPU-GPUs
From the results we can see that:
- Compared running with CPU executable and 12 threads, the jobs run 4.8 times faster with Amber GPU version using one CPU and one GPU.
- Compared running with CPU executable, the jobs run 6.4 times faster with Amber GPU executable with MPI support using two CPUs and two GPUs.
The instructions for submitting jobs the GPU-CPU cluster using GPU executables can be found here.

