MOLECULAR DYNAMICS TO STUDY MUTATIONS IN PROTEIN STRUCTURE
Introduction:
TET2 is found to be a frequently mutated gene in peripheral T cell lymphoma (PTCL) and angioimmunoblastic T cell lymphoma (AITL). TET proteins oxidize modified base – 5-methylcytosine (5mC) on DNA and play important roles in various biological processes. TET2 specifically recognizes CpG dinucleotide and shows substrate preference for 5mC in a CpG context.
Owing to its recently solved crystal structure, more detailed investigation on the structure-function relationship can be explored. The hotspot mutation position – Arg1261 found in AITL, for example, is found to interact with cosubstrate NOG (N-OxalyGlycine) and is critical for its activity; also it may distinguish between normal and modified base. Here, we examine the possible effect of the TET2 mutant on its protein structure and its protein-DNA binding which may shed light on its roles in tumorigenesis.
Molecular dynamics (MD) simulation:
We aim to study the effect of the p.R1261P mutation on the TET2 protein structure and its binding to DNA in due course. We performed a 50ns run of MD simulation in 3 replicates, for both the wild type and mutated form of the protein-DNA complex structure. Generally, we do simulations using two widely used packages: for some analyses we use the GROMACS trajectories whereas for studying changes in binding free energies (DG) we use the program MMPBSA.py in AmberTools12.. The GROMACS simulation was done using the GROMACS 4.5.5 software package with GROMOS96 53a6 force field and AMBER using AMBER12 and amber99SB force field. The starting models for MD simulation of protein TET2 were constructed from the crystal structure and the mutated model.
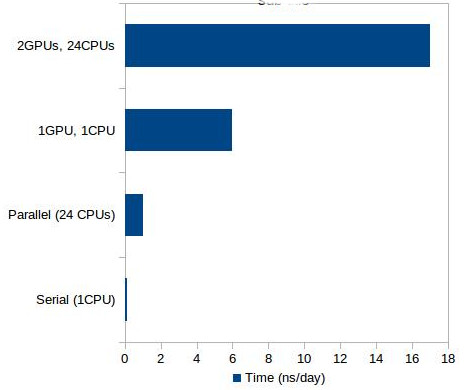
In the present study, we ran the above mentioned simulations using the NUS HPC resources. We ran the GROMACS simulations in the HPC clusters using the parallel executables. The performance was reasonably good at 20ns/day compared to running in a single CPU (in our case with a 4 core CPU, it was 4ns/day). In the case of the AMBER simulations, since it is much slower than GROMACS, the NUS HPC with the recently GPU-enabled resource “goldgpu” was of great advantage.
AMBER GPU acceleration
Presently, the GPU-accelerated MD provides performance that was previously not feasible, even with the help of supercomputers. Today, it has provided a huge boost not only to speed but also significantly reduced energy usage. Furthermore, GPUs can scale well either within a node or over multiple nodes.
In the case of our system, the scale up is seen to be huge – from only 1ns/day using parallel CPU executable to 17ns/day using GPU executable running with 2 GPUs and 24 CPUs. Compared to running serial code in a single CPU, the speed up is more than 100 times. The performance scale-up for our protein system is shown in figure 1.
Results and Discussion:
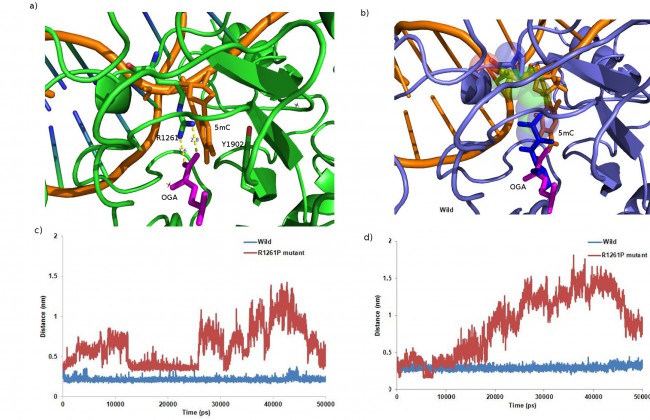
Using molecular modelling and simulation, we analyzed the effect of p.R1261P mutation on the crystal structure of TET2. After modelling and multiple rounds of docking on DNA, there were significant changes found in the intra-atomic interactions within the protein which might affect its function. The mutation hindered the possible cation-p stacking electrostatic interaction seen in the wild type where the modified DNA base (5mC) was sandwiched by an aromatic residue (Tyr1902) and the cation (Arg1261) (figure 2a,b); this was hypothesized to have function in distinguishing modified DNA bases.
The MD simulation of both the wild type and mutated structures were done for 50ns in explicit solvent. Although the total energy of both systems were very similar, the backbone fluctuation was greater in the mutant. The base stacking interaction between Tyr1902 of TET2 and the pyrimidine base of mc6 existed in TET2 throughout the time period of 50ns, but was interrupted in the mutant as Tyr1902 was seen to sufficiently move away from mc6 (figure 2d); the architecture hence proved to be necessary for specific recognition of methyl-CpG dinucleotide by TET2. Also the distance between Arg/Pro1261 (Pro in mutant) and mc6 fluctuated a lot in the mutant (figure 2c).
When checked for hydrogen bonding pattern throughout the simulation, 7 prominent h-bonds were found in the wild type among which 3 were involved with 5mC (modified base), which was seen to be completely lost in the mutated. The binding study showed decreased binding of DNA to the protein in the mutant (decreased free energy of binding, DG) compared to the wild type; this further supported the possibility of loss of function in the mutant. Hence the modelling/docking and the MD simulation data suggested that the mutation might have an altered effect on 5mC-DNA substrate recognition and subsequent protein function.
References:
Chi-Lin Tsai and John A. Tainer. Probing DNA by 2-OG-Dependent Dioxygenase. Probing DNA by 2-OG-Dependent Dioxygenase. Cell 2013; 155:1448-1450.
Hu L, Li Z, Cheng J, Rao Q, Gong W, Liu M, Shi YG, Zhu J, Wang P, Xu Y. Crystal Structure of TET2-DNA Complex: Insight into TET-Mediated 5mC Oxidation. Cell 2013; 155(7):1545-1555.
Plomero et al., Recurrent mutations in epigenetic regulators, RHOA and FYN kinase in peripheral T cell lymphomas. Nature Genetics 2014;46(2):166-170.
Yanagimoto et al., Somatic RHOA mutation in angioimmunoblastic T cell lymphoma. Nature Genetics 2013; 46(2):171-175.

